PDB | 7rjc Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position
Original publication
Member of
copyrightData provided by Research Collaboration for Structural Biology, Brzezinski, P., Whitesell, L., Rubinstein, J.L., Cowen, L.E., Liu, Z., Di Trani, J.M. , provided free of copyright via the Protein DataBank. View original data or read license terms.
Simulation 7rjc_default_dppc
Images
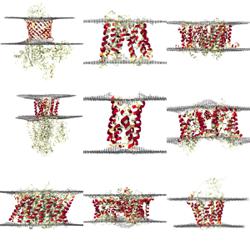
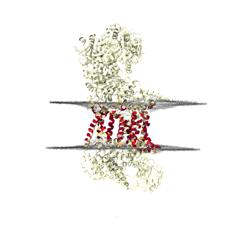
The following pre-rendered images show: distortions of the lipid bilayer, averaged across the entire simulation; positions of lipids in a representative snapshot of the simulation and contacts between lipids or solvent and the protein in either cartoon or surface representation. Rendering is performed using VMD's Tachyon renderer.
Distortions
Distortions show the average surface formed by lipid phosphate beads over the final 800 ns of simulation time. Red indicates a thinning of the bilayer (compared to bulk thickness), whilst blue indicates thickening.
Lipids
Coarse-grained lipids are shown with glycerol beads in yellow, phosphate beads in red and choline beads in blue. Atomistic lipids are coloured according to the CPK standard.
Contacts
Contacts show the average occupancy of the selected group within 6 Å of the protein over the final 800 ns of simulation time. Use the buttons below to select a representation.
Data Download
The following files are available to download and use - hover over to check whether the file is available and click to download. Touch users may need to tap twice to download. Licensed using a Creative Commons Attribution 4.0 International License.
Snapshots
7rjc_default_dppc-ready.pdb
Starting structure used for simulations.
7rjc_default_dppc-atomistic.pdb
Atomistic snapshot created with CG2AT.
7rjc_default_dppc-coarsegrained.pdb
Coarse-grained snapshot (MARTINI representation).
7rjc_default_dppc-distortions.pdb
Distortions (membrane surface representation) in PDB format.
Structures with contacts
7rjc_default_dppc-head-contacts.pdb
Atomistic structure with headgroup contacts as temperature factors.
7rjc_default_dppc-tail-contacts.pdb
Atomistic structure with acyl tail contacts as temperature factors.
7rjc_default_dppc-solvent-contacts.pdb
Atomistic structure with solvent contacts as temperature factors.
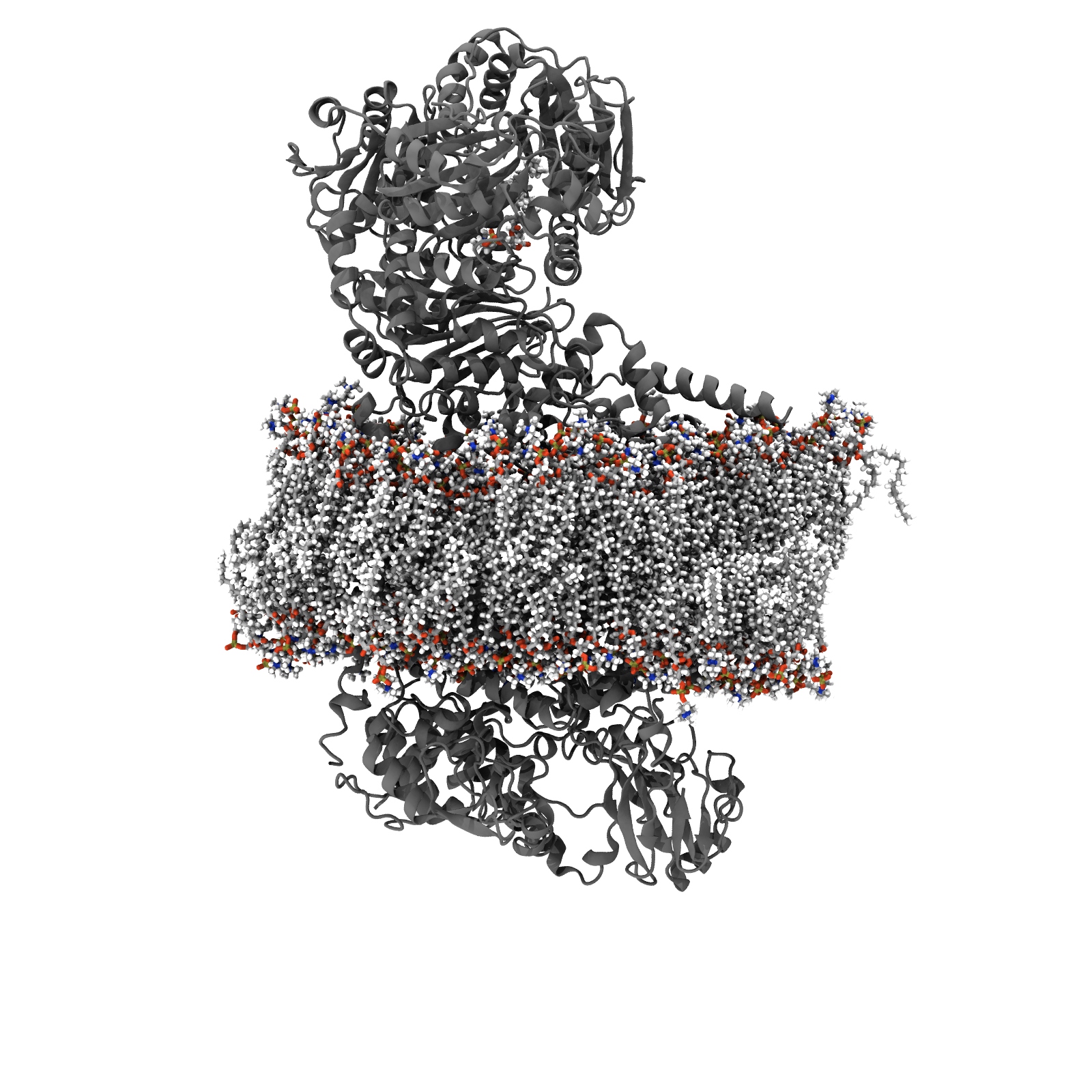
View molecule in 3D
View an atomistic snapshot from this simulation. You will not need to install any additional software or plugins.
check Your browser is ready to display 3D content using WebGL.
checkA structure is available through the MemProtMD API.
errorThis is an extremely large file - loading it may temporarily cause serious performance issues.
insert_drive_file
7rjc_default_dppc.mpmd.finalframe.atomistic.pdb
9.2 MB, PDB Format
autorenewLoading structure
Show protein as:
Colour protein by:
Show lipids as:
Colour lipids by:
Polypeptide chains
Select a chain to view more detailed analysis. The cards below show a vertical violin plot for each chain, with shapes representing the normalised density of all residues (white), alpha helical residues (blue) and beta sheet residues (green) along the bilayer normal.